CSOmap
My first project in computational Biology
This is my undergraduate work at Dr. Zemin Zhang’s Lab at Peking Unviversity.
It starts with an interesting question: Can we infer the spatial information from scRNA-seq that is usually lost during tissue dissociation? We made a simple but reasonable assumption that spatially-close cells should have high ligand-receptor interactions. Based on that we developed CSOmap, a computational tool to infer cellular interaction de novo from scRNA-seq. We show that CSOmap can successfully recapitulate the spatial organization of multiple organs of human and mouse including tumor microenvironments for multiple cancers in pseudo-space, and reveal molecular determinants of cellular interactions. Further, CSOmap readily simulates perturbation of genes or cell types to gain novel biological insights, especially into how immune cells interact in the tumor microenvironment. CSOmap can be a widely applicable tool to interrogate cellular organizations based on scRNA-seq data for various tissues in diverse systems.
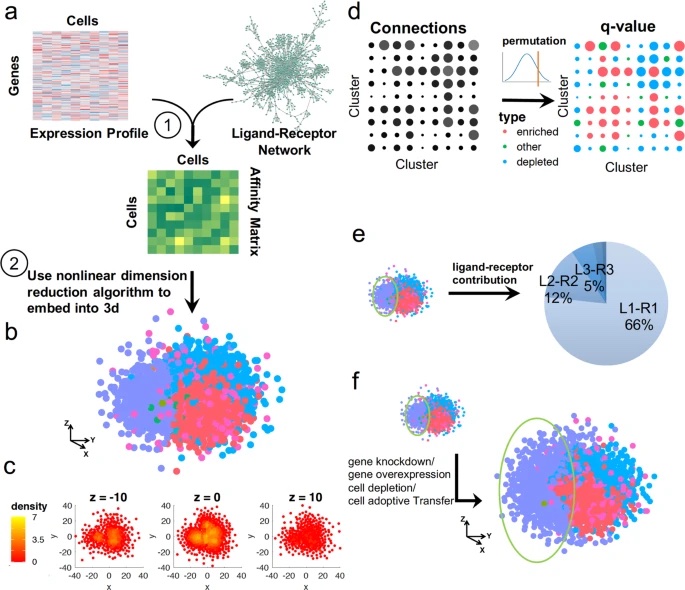
It is a cool project, I really appreciate the mentorship from Dr. Xianwen Ren and Dr. Zemin Zhang. I’ll never forget this experience.
For details, please check our manuscript and GitHub repository.